Malaria Cell Image Classification
Date: 06 Nov 2021 Tag(s): Jupyter Notebook, Python, Pandas, FastAI, Malaria, Cells, Images, CNN, Tensorflow, Keras Categories: Machine Learning, ML, Classification, Neural Network Download Project: 10.9 MB - Zip ArchiveOverview #
Malaria is a life-threatening disease caused by parasites that are transmitted to people through the bites of infected female mosquitoes, it is preventable and curable. Malaria is specifically caused by parasites that are spread to people through the bites of infected female mosquitoes. This project aimed to create a model that would be able to identify cells that are either infected or uninfected with malaria. In my approach I decided to create image classification models using two different libraries: FastAI and Tensorflow/Keras.
NOTE: The total number of images in this dataset was 27,560 (13,780 in each folder: Uninfected and Parasitized). Due to the technical limitations of my machine, I had to subset the dataset and use 1/4th (6,890) of the total images (3,445 in each folder) in order to complete the modeling in a reasonable amount of time.
This analysis has two parts:
This notebook builds two different (FastAI and Tensorflow/Keras) CNN (Convolutional Neural Network) image classification models for identifying cells that are either infected or uninfected with malaria.
View Jupyter Notebook #
The notebook goes step-by-step through the project, please follow the directions and cell order if you would like to replicate the results.
- Click the "View Notebook" button to open the rendered notebook in a new tab
- Click the "GitHub" button to view the project in the GitHub portfolio repo
Part 1 - FastAI #
FastAI is a modeling library built on top of PyTorch and is quite straight forward to use.
Augmented Images #
After reducing the size of the dataset, the first step is to normalize their size (make all images the same size) and slightly augment some of them (change shape, blur, rotate, etc…) because in a real world environment the model will not always have perfect images to work with.
Below is a small batch of those augmented images:
Model Training #
Model was build using a built-in model trainer of FastAI, XResNet34, and ran for 5 epochs, below are the results for each epoch:
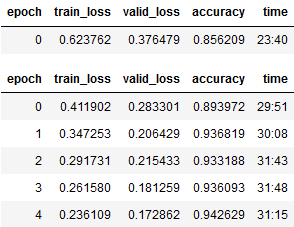
NOTE: The stand-alone epoch at the top is the first run, base-line, of the model. The remaining epochs are fine-tuned from this initial run.
The final fine-tuned model ends up with an accuracy of ~0.94, with a loss of 0.17, after ~3 hrs of runtime. Very good
results, especially considering the massively reduced amount of images I had to use for modeling.
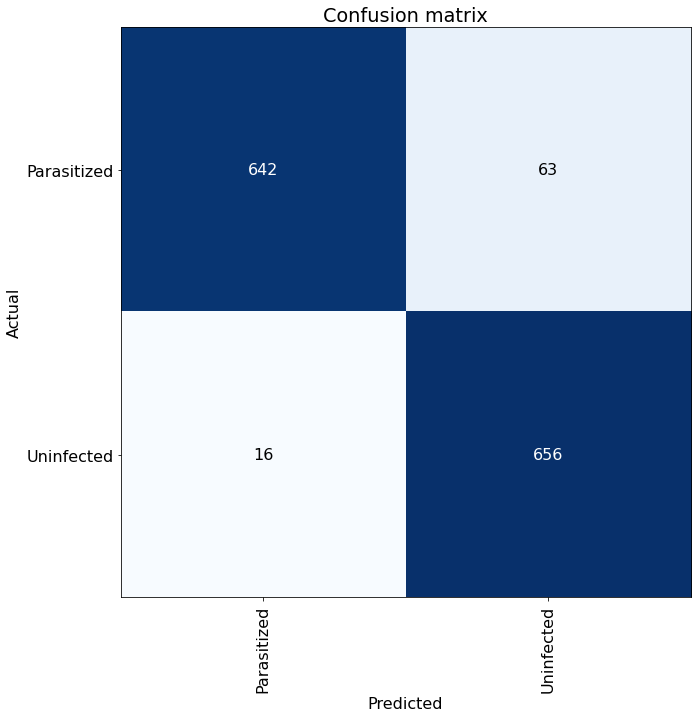
Confusion Matrix #
From the matrix below, we can see that the model predictions were very good and made relatively few mistakes. The model
incorrectly identified 63 Uninfected cells as Parasitized and 16 Parasitized cells as Uninfected from this sampling.
Classification Report #
The classification report just gives and overview of how the model performed with various classification metrics: precision, recall and f1-score.
precision recall f1-score
Uninfected 0.94 0.97 0.95
Parasitized 0.97 0.94 0.95
accuracy 0.95
macro avg 0.95 0.95 0.95
weighted avg 0.95 0.95 0.95
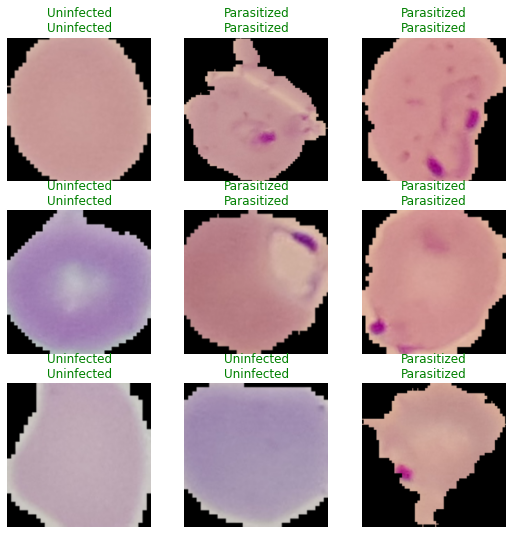
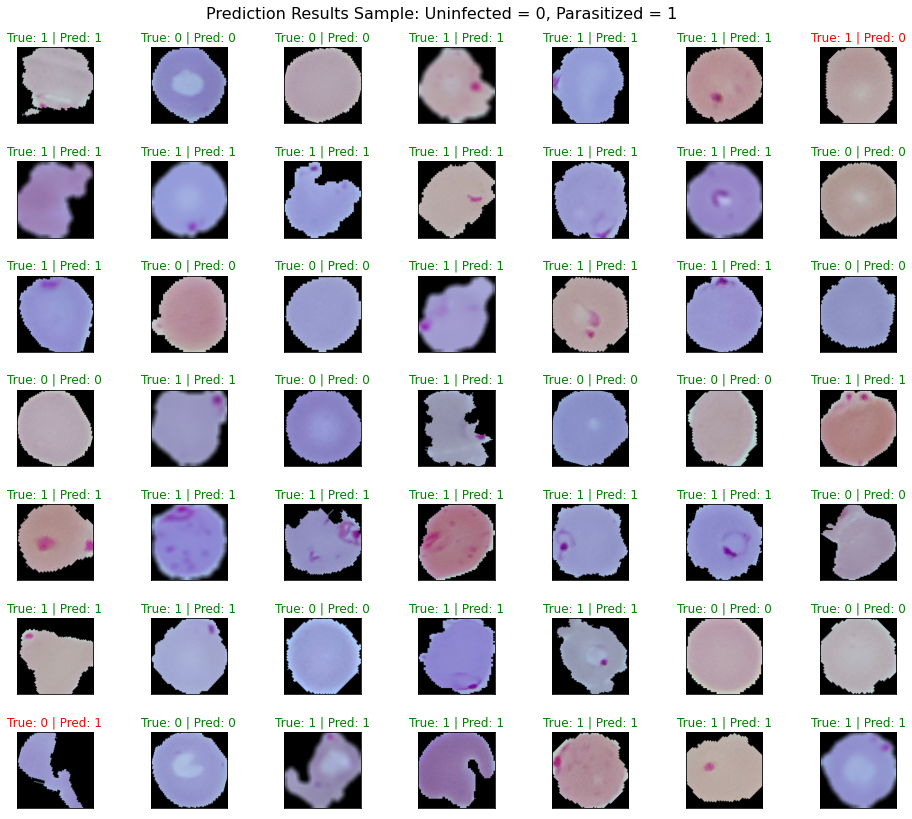
Model Predictions and Conclusion #
Even though I had to severely reduce the number of images for training, the model still performed very well with an
accuracy score of ~0.94. The length of the modeling (~3 hrs) for the amount of images is far longer than I would have
liked but my computer is currently just not up to spec, having a more powerful GPU and CUDA
installed will greatly speed up the modeling process. FastAI definitely makes it very easy to use and does a lot of the
groundwork for you, I am definitely quite please with the results and the library as a whole.
Now we can move onto to making a CNN model from scratch using Tensorflow/Keras and see how it compares to the FastAI model.
Part 2 - Tensorflow/Keras #
Keras is a modeling library built on top of the very popular (if not the most popular) python machine learning library: Tensorflow.
Augmented Images #
Like before, in the FastAI section, the dataset was reduced and the images were augmented, below is a sample of those images:
Model Training #
This time I will not be able to use a built-in model to bounce off of like in FastAI and instead will need to build a model myself from scratch.
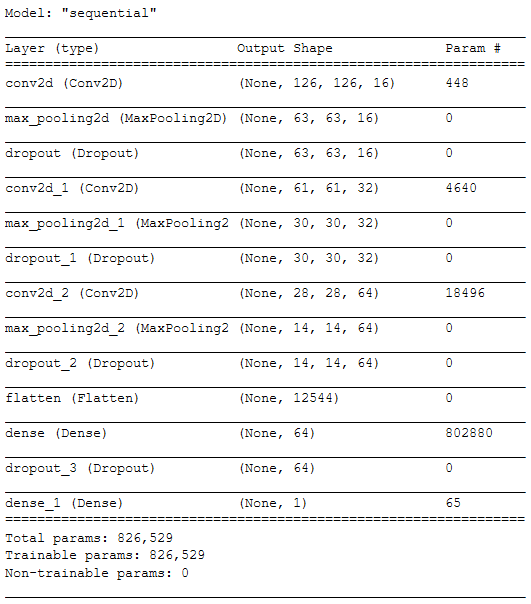
Model Layers and Parameters #
Below are the layers and parameters for the model:
Model Results - Accuracy and Loss #
The model was slated to run for 20 epochs but I implemented an early stop loss into the model fitting, thus it only ran for 12 epochs instead.
The modeling ended with an accuracy score of ~0.9620 after 12 epochs and ~1.5 hrs of runtime (half the runtime of the FastAI model),
which is very good and just slightly better than the FastAI modeling. The modeling also ended early, at epoch 12/20, as
val_loss started to rise again, indicating that the peak number of epochs had been reached and any further training would not be beneficial.
Accuracy Graph #

Loss Graph #
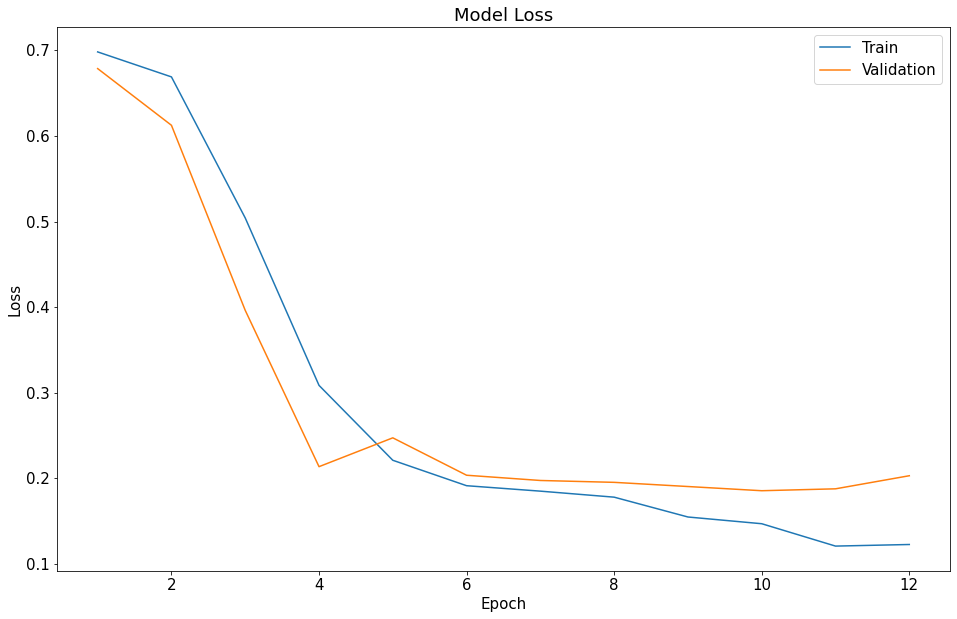
NOTE: You can see the orange “Validation” line uptick slightly at epoch 12, which resulted in model training stopping early.
Confusion Matrix #
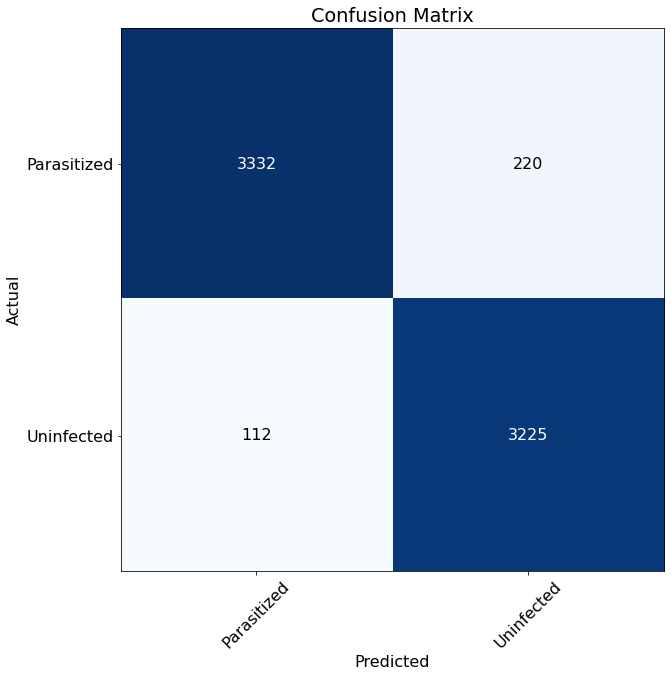
From the matrix below, we can see that the model predictions were very good and made relatively few mistakes. The model
incorrectly identified 220 Uninfected cells as Parasitized and 112 Parasitized cells as Uninfected from the sample.
Classification Report #
The classification report just gives and overview of how the model performed with various classification metrics: precision, recall and f1-score.
precision recall f1-score
Uninfected 0.94 0.97 0.95
Parasitized 0.97 0.94 0.95
accuracy 0.95
macro avg 0.95 0.95 0.95
weighted avg 0.95 0.95 0.95
Model Predictions and Conclusion #
See Part 1 - Model Predictions and Conclusion for FastAI results.
Again, even with the severely reduced number of images for training, the model performed very well with an accuracy
score of ~0.96. The Tensorflow/Keras modeling (about 1.5 hrs) took about half as long as the FastAI modeling (~3 hrs)
to complete, which is a great improvement in that regard. In terms of model performance, the keras model performed just
slightly better than the FastAI model (~0.94). Given that they are so close to each other, I’m willing to chalk it up
to margin of error and say that they perform about the same. As a result, this leaves the model training time as the
deciding factor for the better model.
Overall Conclusion #
Both of these CNN model implementations have their pros and cons, FastAI makes it very simple to create, run, sample, and predict with very little extra work on the users part. The drawback to this is that you don’t have as much finer control of your model and largely left to the whims of the library. Tensorflow/Keras however, provides much finer control over pretty much all aspects of the modeling process but at the cost of having to create it all yourself, thus longer time for development. It’s hard to say which of these models is better but I think it largely depends on the project constraints and how much time you are willing to spend on it.
My opinion is:
- If you just want to get to the modeling with minimal fuss/building and have most everything built-in for you, I would recommend using FastAI.
- If you want to build everything yourself and have much finer control over your modeling, I would recommend using Tensorflow/Keras.
In either case, I can’t recommend enough how much better your modeling life will be with a better GPU and CUDA, especially so if you are doing image processing.